Biology
Part I: BioChemistry and Microbiology
1. Describe the basic structure of proteins, lipids, phospholipids, carbohydrates and nucleic acids and the basic chemical bonds and forces that hold their respective monomers together.
a) Write a general structure for an amino acid?
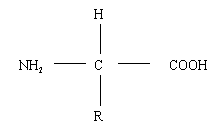
General structure for an amino acid.
b) What is the function of the R group of amino acids?
The R group determines the chemical bonding ability of the amino acid - depending on the different R groups, the amino acid may be able to participate in hydrogen bonding, hydrophobic interactions, salt bonds, or disulphide bridging.
c) Draw the structure of the peptide bond. What type of polymer is held
together with peptide bonds?
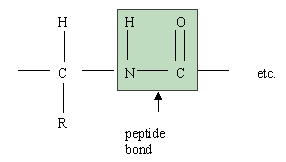
Proteins
d) Proteins have primary, secondary, tertiary and quaternary structure. Name the chemical bond types or forces that hold each of these structures together.
Primary structure of proteins is held together by covalent peptide bonds.
Secondary structure (e.g. alpha helix, B-sheet, or random coil) is held together by hydrogen bonding.
Tertiary structure (the overall folded appearance of the protein) is held together by hydrogen bonding, hydrophobic interactions, salt bridges, and disulphide bridges.
Quaternary structure (the organization of multi-subunit proteins) is held together by hydrogen bonding, hydrophobic interactions, salt bridges and disulphide bridging.
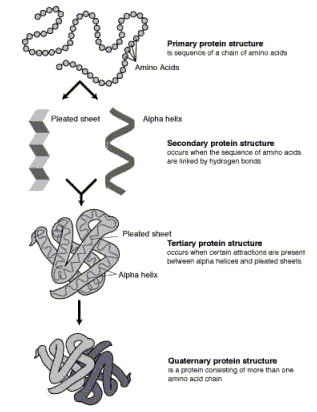
illustration by Darryl Leja, National Human Genome Research Institute http://www.nhgri.nih.gov/DIR/VIP/
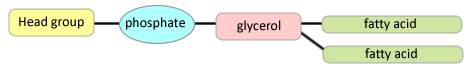
e) What properties of lipids make them able to form membranes?
The amphipathic character of the lipids allows them to form membranes. That is, these molecules have one polar end and one hydrophobic end. Thus they self-assemble into layers with the hydrophobic portions together and the polar areas together.
The hydrophobic portions want to be away from water while the polar portions want to be in contact with water. This results in a bilayer with a hydrophobic center.
f) Draw a typical phospholipid.
g) What is the role of calcium ions in the construction of membranes?
Calcium ions provide bridging between the negative charges of the phosphate groups to prevent electrostatic repulsion from making the membranes leaky.
h) Name the two types of nucleic acids found in cells, and describe their differences and similarities.
The two types of nucleic acids are DNA (deoxyribonucleic acid) and RNA
(ribonucleic acid). They are similar in that both are information carrying polymers (as opposed to structural polymers such as collagen), both use the bases A (adenosine), C (cytosine) and G (guanosine),
both have backbone structures consisting of alternating units of 5
carbon sugars and phosphate, and both can be single or double stranded depending on the situation, and both are assembled by enzymes.
Differences include: DNA is usually double stranded, it uses T (thymine) as the fourth base, the sugar for the backbone is deoxyribose, and it stores genetic information (replication is more stringent than for RNA). RNA translates genetic information into structures such as proteins, uses U (uracil) as the fourth base, and the sugar for the backbone is ribose.
i) What type of chemical bonding holds DNA together?
Phosphodiester bonds hold DNA together.
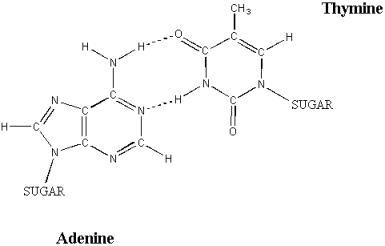
j) Why can C only pair with G, and A only pair with T?
This is due to the limited space within the DNA double helix (where the bases sit) and the need to maximize hydrogen bonding potential (which in turn maximizes the stability of the DNA helix). Two purines (e.g. A and G) together would be too large to be accommodated in the available space (the sides of the helix would bulge). Two pyrimidines (e.g. C and T) would be too small (the bases wouldn't be within hydrogen bonding distance). Purine-pyrimidine pairs are the correct size for the observed diameter of the double helix. A and T paired together are in the correct position to allow the formation of two hydrogen bond pairs, while C and G paired together allow the formation of three hydrogen bond pairs.
An AT base pair. Note the two hydrogen bonds holding the bases in place (hydrogen bonds are shown by dotted lines).
illustration from: www.ncc.gmu.edu/dna/two.htm
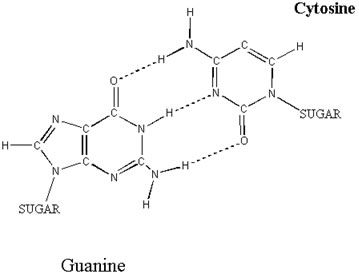
A GC base pair. The three hydrogen bonds holding the bases together make this a more sturdy pairing than the AT pair shown above (hydrogen bonds are shown as dotted lines). illustration from: www.ncc.gmu.edu/dna/three.htm
k) What is the basic chemical formula for a carbohydrate?
The basic chemical formula for a carbohydrate is (CH2O)n.
l) Name a six carbon sugar.
Six carbon sugars include glucose, fructose, galactose, etc.
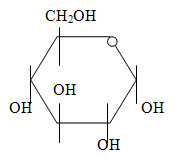
m) Draw the structure of glucose in the closed conformation.
n) Draw a glycosidic bond.
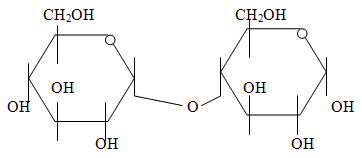
A glycosidic bond is a bond formed between two sugar monomers by a dehydration reaction (with the loss of water). The example shown above is a 1,4 glycosidic bond between two monomers of glucose.
2. Describe the role of the above polymers in the cell.
a) What is the role of enzymes in the cell? Give an example of an enzyme.
Enzymes increase the rate of chemical reactions in the cell by lowering the activation energy necessary for the reaction to occur. Enzyme names usually end in the suffix "ase". An example of an enzyme is amylase, a protein which breaks down starch.
b) Why don't ionized molecules pass readily through the plasma membrane?
Ionized molecules do not pass readily through the plasma membrane because it requires a very large amount of energy to move charged molecules through the hydrophobic core of the plasma membrane. The energy needed to disrupt the hydrophobic bonds between the fatty acid tails of the phospholipids is not spontaneously available.
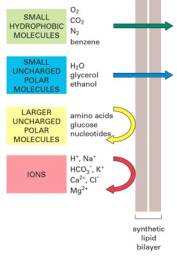
illustration from: http://www.accessexcellence.com/AB/GG/memPerm.html
c) Distinguish between the roles of rRNA, tRNA and mRNA within the cell.
Ribosomal RNA (rRNA) makes up the scaffolding for the ribosome. All ribosomal proteins bind to either the rRNAs or to proteins bound to the rRNAs.
Messenger RNA (mRNA) carries instructions for building proteins from the DNA to the ribosomes. Acts as a template for protein synthesis.
Transport RNA (tRNA) moves amino acids to the ribosome. This folded molecule has two distinct ends. One end which attaches to the amino acid it carries; the other which is the anticodon which base pairs with the codon on the mRNA to ensure that the correct amino acid is added to the growing peptide chain.
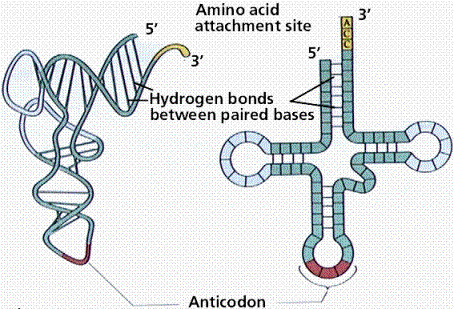
illustration from: http://gened.emc.maricopa.edu/Bio/BIO181/BIOBK/tRNA2.gif
d) Name three carbohydrate based structures found in prokaryotic cells.
Three carbohydrate based structures in prokaryotes include: capsules, slime layers, lipopolysaccharide (outer membrane), peptidoglycan, storage granules of poly-β-hydroxybutyrate or glycogen.
e) What is the role of DNA in the cell?
DNA acts as a genetic information storage center.
f) How can prokaryotic and eukaryotic DNA be distinguished at the molecular level?
Prokaryotic DNA is a single circular chromosome, and it is compacted by histone-like proteins. Eukaryotic DNA consists of many linear chromosomes which are compacted by histones.
3. Clearly distinguish between prokaryotic and eukaryotic cells.
a) Name three ways that prokaryotic and eukaryotic cells are structurally different.
Prokaryotes |
Eukaryotes |
- have peptidoglycan
- no nucleus
- no sterols in the membranes
- no membrane bounded organelles
- no mitochondria
- no chloroplasts
- much smaller
- no internal membrane systems
- single, covalently closed chromosome
|
- cell walls may be made of pectins, cellulose, silica, etc.
- sterols (e.g. cholesterol) used to maintain membrane fluidity
- nucleus present
- membrane bounded organelles (including mitochondria and chloroplasts) present
- extensive internal membrane systems much larger
- multiple, linear chromosomes present
|
b) Do prokaryotic cells undergo meiosis? Why or why not?
Prokaryotes do not undergo meiosis, they multiply by binary fission. During this process, their single chromosome becomes attached to the membrane at two points which move apart, ensuring that each daughter cell receives one copy of the chromosome. Note that prokaryotes are already
in cells, and have no need for a reductional division to produce gametes. They do not reproduce sexually except to a limited extent (F plasmids), which rarely result in more than a partial diploid state for the recipient cells.
c) What eukaryotic organelles are thought to have arisen from prokaryotic cells? Describe the evidence available for this theory.
The organelles in question are the mitochondria and chloroplasts. Evidence for the endosymbiont theory includes:
- Both organelles are approximately the same size as an average bacterium.
- Both organelles reproduce by binary fission (as bacteria do).
- Both organelles have their own closed circular DNA.
- Both organelles have their own 70S ribosomes (same size and function as for bacterial ribosomes - eukaryotic ribosomes are larger - 80S) and their own tRNA.
- There are currently examples of endosymbiotic relationships which may be at an earlier stage in their association (i.e. not yet as intimately involved as eukaryotic cells and their chloroplasts or mitochondria).
4. Describe the construction of the prokaryotic peptidoglycan, and its role in the cell.
a) Name the two sugars that form the backbone of peptidoglycan.
N-acetylglucosamine and N-acetylmuramic acid are the sugars which make up the peptidoglycan backbone.
b) What does peptidoglycan do for the cell?
Peptidoglycan determines the cell shape and supports the plasma membrane against osmotic pressure when the cell is growing in a hypotonic environment relative to the internal conditions in the cell.
c) Both lysozyme and penicillin cause the peptidoglycan to be broken down. Describe the mechanism by which each of these agents causes cell lysis.
Lysozyme is an enzyme which cuts the glycosidic bonds of the sugar chains of peptidoglycan between the N-acetylmuramic acid and the N-acetylglucosamine. This weakens the net-like structure of the peptidoglycan, and allows the cell to lyse due to osmotic pressure.
Penicillin is an antibiotic which acts a molecular mimic for the peptide portion of the peptidoglycan subunits. It binds irreversibly into the active site of the penicillin binding proteins (a family of proteins which are responsible for polymerizing peptidoglycan) and prevents peptidoglycan synthesis. It also binds to the active site of the transpeptidation enzymes which crosslink the peptidoglycan chains. This prevents the crosslinking of the chains and further weakens the peptidoglycan net. The penicillin also binds to the peptidoglycan hydrolases. These enzymes normally function to make small nicks in the peptidoglycan to allow the insertion of new subunits so that the cell wall can grow as the cell gets larger. Penicillin makes these enzymes more active, resulting in many nicks in the already established peptidoglycan, but there are no new subunits being inserted (Penicillin binding proteins are not functioning). Thus the peptidoglycan net becomes unstable, and eventually the cell lyses due to osmotic pressure.
d) Describe the assembly of peptidoglycan in the cell.
Peptidoglycan is assembled in three distinct parts. In the first part, soluble enzymes link the N-acetylglucosamine (glcNAc) and N-acetylmuramic acid (murNAc). The peptide is then assembled enzymatically onto the murNAc part of the subunit. That is, each amino acid of the peptide is added by a separate enzyme, rather than having the entire peptide assembled on a ribosome and then attached to the disaccharide. The second part of the process involves moving the assembled subunits across the plasma membrane. The subunits are carried across the membrane by a carrier lipid (undecaprenol phosphate), and released on the other side of the membrane. The final part of polymerization involves the polymerization of the subunits into short chains and their insertion into the peptidoglycan net by the penicillin binding proteins. Once in place, the new chains are crosslinked to older parts of the net by the transpeptidases.
e) Discuss how peptidoglycan lends strength to the cell envelope.
Peptidoglycan is a large net like structure in which individual chains are covalently linked to chains on either side or in the layers above and below. The extensive crosslinking between chains and layers makes this polymer strong and stretchy. It supports the cell envelope because it limits how far the plasma membrane can stretch due to osmotic pressure by providing mechanical support to the membrane.
5. Differentiate between Gram positive and Gram negative cells on the molecular level.
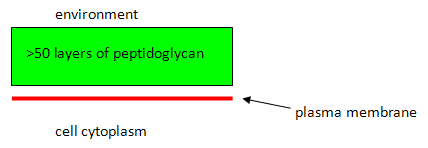
a) Draw a labeled diagram of the Gram positive cell envelope.
b) In the Gram stain, do you expect the Gram positive cell to retain the primary or the secondary stain?
You expect the Gram positive cell to retain the primary stain
(purple) - the peptidoglycan will dehydrate during the selective decolourizing step (acetone alcohol rinse) which will trap the primary stain within the cell.
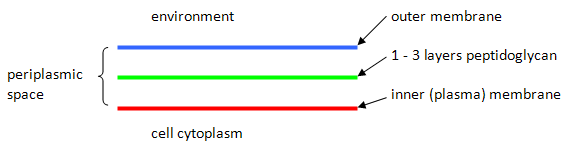
c) Draw a labelled diagram of the Gram negative cell envelope.
d) Following the Gram stain procedure, what colour do you expect Gram negative cells to be? Why?
You expect Gram negative cells to be red. During the decolourizing step, the acetone alcohol will act as a dehydrating agent to shrink the pores in the peptidoglycan and as a lipid solvent to dissolve portions of the membranes and to mobilize the primary stain. Because the peptidoglycan is so thin in Gram negative cells as compared to Gram positive cells, the dehydrating action is unable to trap the dye, which is lost in Gram negative cells during this step. Thus, the Gram negative cells have many available binding sites for the secondary stain, which is red.
e) Describe the outer membrane of Gram negative cells at the molecular level. What components are unique to this membrane?
The outer membrane of Gram negative cells can be separated into an inner and outer leaflet. The outer leaflet is made up primarily of lipopolysaccharide,(a polymer unique to the outer membrane). The inner leaflet is made up mostly of phospholipids. The entire membrane contains many embedded proteins which are responsible for transporting or allowing movement of various substances across the outer membrane. Many of these proteins are part of the porin family.
6. Describe how molecules are transported across membranes.
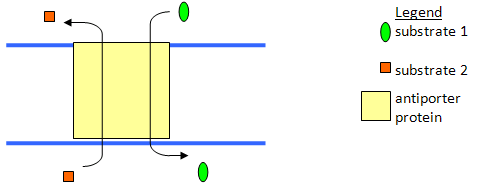
a) What is a symporter?
A transport protein which moves two soluble substrates across a membrane at the same time in the same direction. The energy required is generated by at least one of the substrates moving down its concentration gradient as it crosses the membrane. The other substrate is usually being concentrated against its concentration gradient.
b) Diagram an antiporter?
c) What are the features of a membrane bound protein that make it different from a cytoplasmic protein?
There are two main ways in which a protein can be membrane bound: it may have a hydrophobic anchor or it may have a hydrophobic surface. If the protein has a hydrophobic surface, it may become bound into the structure of the membrane. In many cases, the protein will have a pore or channel in its center which is hydrophilic. Alternatively, the protein may have a covalently attached fatty acid or phospholipid which will become associated with a membrane, and the protein then becomes membrane bound. If the hydrophobic anchor is removed, the protein usually is quite water soluble.
d) What is a porin, and where is it found in the cell.
A porin is a hydrophobic barrel shaped protein with a hydrophilic channel passing through its center. It is a membrane protein found in the outer membranes of Gram negative bacteria and mitochondria which allows the nonspecific passage of hydrophilic molecules through the membrane. All the molecules must be small enough to pass through the central pore and will be moving down their concentration gradient. This is a form of passive transport.
e) Describe two ways that a cell can concentrate molecules inside the cell against a concentration gradient.
There are several options for moving molecules across membranes against their concentration gradients:
- binding protein systems using ATP as the energy source (active transport).
The solute and ATP will bind to the membrane transport protein, the ATP will be hydrolyzed, and the energy released will allow the transport of the solute across the membrane.
- active transport using the proton motive force as the energy source.
May use a symporter (direct use of the proton motive force) - the proton will enter the cell coming down its concentration gradient and this energy will drive the importation of the second solute.
Alternatively, the proton motive force may be used to generate a second chemical gradient (e.g. Na+) which can then be harnessed in an antiporter to import the second solute while exporting Na+ ions.
- group translocation: the substance being imported is modified during transport to maintain an apparent concentration gradient which is higher outside than inside the cell. For example, glucose can be imported into the cell down its concentration gradient (and therefore "under its own power") by phosphorylating it during transport. Thus a high concentration of glucose-phosphate is being generated inside the cell, but the concentration of glucose inside the cell is very low. As a result, glucose will continue to drive its own entry into the cell.
7. Outline the steps involved in the generation of a protein from the gene sequence found in DNA.
a) Write the sequence you would expect to find for the mRNA given the following DNA sequence:
ATGCCATGTATTGCGCAA.
The expected sequence is: UAC GGU ACA UAA CGC GUU
b) How many codons are there in the sequence shown in (a)?
There are six codons in the sequence.
c) Describe the structure of a ribosome.
Prokaryotic and eukaryotic ribosomes are similar in function and form, but differ slightly in size. Both are made up of a large and a small subunit. The assembled prokaryotic ribosome is a 70S structure made up of a 30S and a 50S subunit. The small subunit has a 16S rRNA and 21 proteins, while the large subunit has a 5S and a 23S rRNA as well as 34 proteins.
The eukaryotic ribosome is an 80S structure made up of a 40S small subunit (18S rRNA, 33 proteins) and a 60S large subunit (5S, 28S, and 5.8S rRNAs, 49 proteins).
d) How does a ribosome make a protein?
- initiation: mRNA binds to the small ribosomal subunit, and the initiator tRNA binds to the initiator codon.
- the large ribosomal subunit binds to the above complex.
- The ribosome has 2 active sites, the A (aminoacyl) site and the P (peptidyl)site. The initiator tRNA is in the P site. An aminoacyl tRNA will recognize the mRNA codon which is present in the A site.
- A peptide bond forms between the amino acids on the two tRNAs (the growing chain is transferred onto the tRNA in the A site).
- The codons translocate one position down to the P site
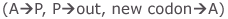
- Repeat until the stop codon arrives in the A site.
e) What is a tRNA?
A tRNA (transfer RNA) is a single stranded RNA, which moves amino acids into the correct position on the ribosome for incorporation into growing proteins. These molecules are L-shaped when folded. One end of the L is enzymatically linked to an amino acid, the other end is the anticodon that binds to the codon sequence of the mRNA.
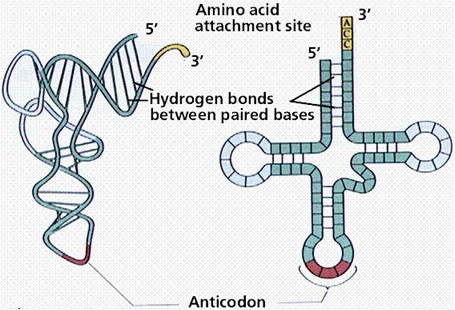
illustration from: http://gened.emc.maricopa.edu/Bio/BIO181/BIOBK/tRNA2.gif
f) What is RNA polymerase, and what does it do in the cell?
An RNA polymerase is an enzyme which combines ribonucleotides into an RNA chain. These enzymes are DNA dependent - that is, they require a DNA molecule to act as a template. These enzymes make all the types of RNA present in the cell.
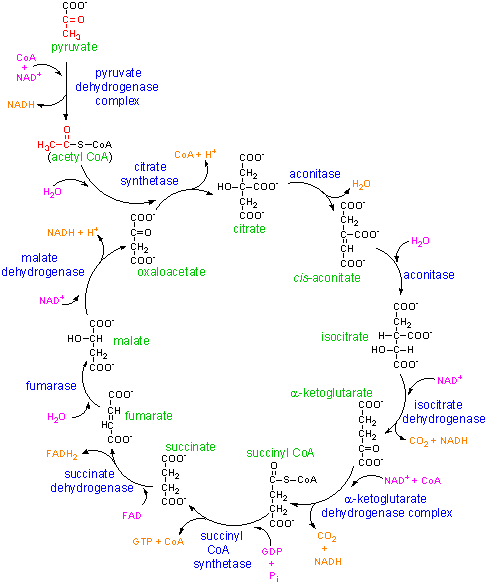
8. Outline the reactions involved in glycolysis, the TCA cycle (Kreb's cycle) and the electron transport for aerobic metabolism.
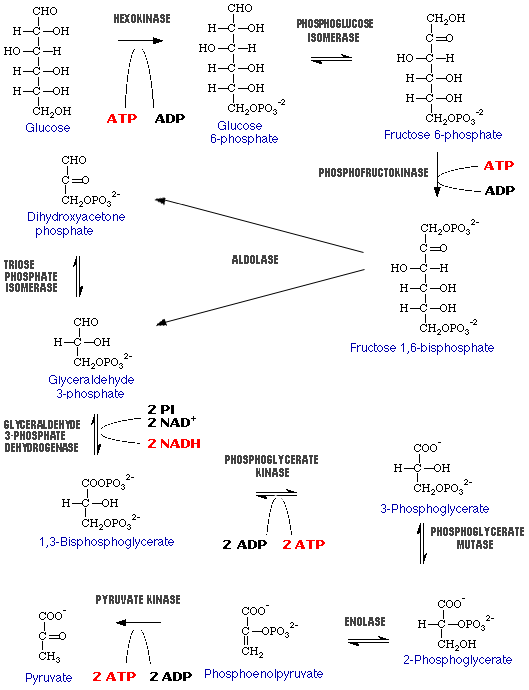
a) Draw the reactions involved in glycolysis.
b) How many net ATP do you gain from glycolysis?
You generate a net 2 ATP.
c) What is the final product generated by glycolysis?
The final product is 2 molecules of pyruvate.
d) Draw the reactions of the TCA or Kreb's cycle.
e) What are the products of the TCA cycle?
The products of the TCA cycle are 2 CO2, 1 GTP, 3 NADH, 1FADH2 per turn of the cycle. (Note: cycle turns twice for 1 glucose molecule entering glycolysis.)
f) How many ATP do you generate for each turn of the cycle? (Assuming that all NADH and FADH2 are put through the electron transport chain).
The total per turn of the cycle is 1 ATP (from the GTP), 9 ATP (generated from the 3 NADH), and 2 ATP (generated from the FADH2) for a grand total of 12 ATP per turn of the citric acid cycle.
g) Sketch the electron transport chain.
This is the first part of the electron transport chain. The NADH reducing equivalents (2 electrons and 1 proton)and a proton from the bulk solvent in the cell are passed to the NADH coenzyme Q reductase. This enzyme complex will eject the protons to the intermembrane space and pass the electron to ubiquinon (a mobile electron carrier). The Ubiquinon will pass the electron to cytochrome reductase enzyme which will pick up a proton from inside the cell to balance the charge of the electron. Cytochrome reductase will pass the electrons to cytochrome C and will eject the proton to the outside of the cell.
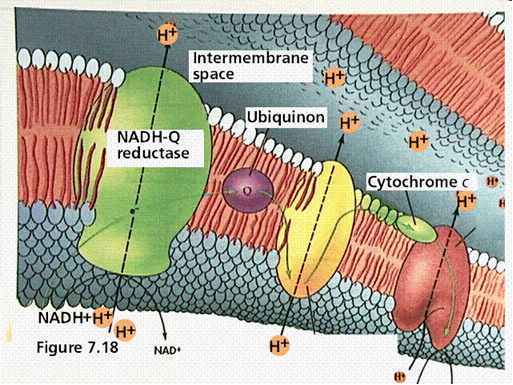
illustration from: http://gened.emc.maricopa.edu/Bio/BIO181/BIOBK/chemiosm1.gif
In the second part of the electron transport chain, cytochrome C will pass the electrons to cytochrome oxidase
which picks up a proton from inside the cell. The enzyme splits oxygen
and combines it with two protons. Another proton is ejected across the membrane in the process. This
results in a build up of protons on the outside of the membrane, which causes a charge differential to form known as the proton motive force. This proton motive force can be used to generate ATP via the ATP synthase enzyme complex. Protons are allowed to flow down their concentration gradient into the cell, and the enzyme harnesses this energy to convert ADP and inorganic phosphate into ATP.
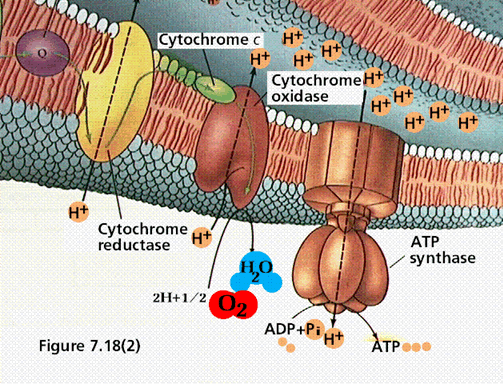
illustration from: http://gened.emc.maricopa.edu/Bio/BIO181/BIOBK/chemiosm2.gif
h) What is the purpose of the electron transport chain?
The purpose of the electron transport chain is to recycle the reducing equivalents (NADH, and FADH2) generated during glycolysis and Kreb's cycle to NAD+ and FAD, to create the proton motive force and conserve the bond energy of the sugar.
i) What does the cell do with the electrochemical gradient generated by the electron transport chain?
The cell can use this gradient to generate ATP, to move, to transport substrates (use the proton gradient, either directly or indirectly), etc.
j) How many ATP can be generated for each molecule of glucose by aerobic respiration?
Thirty eight molecules of ATP can be generated per glucose molecule by aerobic respiration.
9. Describe anaerobic respiration and fermentation, and how they are different from aerobic metabolism.
a) Does anaerobic respiration generate as much energy as aerobic metabolism?
No, because the alternative electron acceptors (e.g. nitrate, sulphate, etc) have a lower reduction potential than does oxygen. That is, the difference between the donor NADH and the inorganic acceptor molecule is less than between NADH and oxygen. The energy yield is related to the size of the reduction potential - a smaller energy yield allows less ATP to be made.
b) Define anaerobic respiration.
An anaerobic energy yielding process in which the final electron acceptor from the electron transport chain is an inorganic molecule other than oxygen.
c) Give two examples of products of fermentation which are sold commercially.
Examples include beer, wine, cheese, antibiotics, amino acids, yogurt, sour cream, etc.
d) Define fermentation.
An energy yielding process in which organic molecules act as both the electron donor and the electron acceptor.
For example: glucose may be converted to pyruvate by glycolysis (glucose donates the electrons to NADH). Pyruvate can then act as the electron acceptor to regenerate NAD+ and pyruvate is converted to lactic acid.
Note: this is a relatively inefficient way to make energy, so for organisms which do not rely exclusively on fermentation, it will not occur in the presence of oxygen.
e) Name three alternative electron acceptors for anaerobic respiration.
Three alternative electron acceptors for anaerobic respiration include nitrate, sulphate, carbon dioxide, elemental sulphur, and ferric iron.
f) What is a facultative anaerobe?
A microorganism which does not require oxygen for growth, but that does grow better in its presence.
g) Define microaerophile.
A microorganism that requires low levels of oxygen for growth, around 2 to 10%, but is damaged by normal atmospheric levels of oxygen.
h) Differentiate between homolactic and heterolactic fermentation.
A homolactic fermentation takes sugars and generates lactic acid as the sole fermentation product (e.g. lactic acid bacteria used for making yogurt).
A heterolactic fermentation takes sugars and generates a mixture of lactic acid, ethanol and CO2.(e.g. some yeasts used in the brewing industry)
i) Give three examples of the biochemical products of mixed acid fermentation.
Examples of the biochemical products of mixed acid fermentations include ethanol, acetic acid, formic acid, lactic acid, succinic acid, carbon dioxide, hydrogen gas and butanediol.